Taming your datasets with MeDUSA
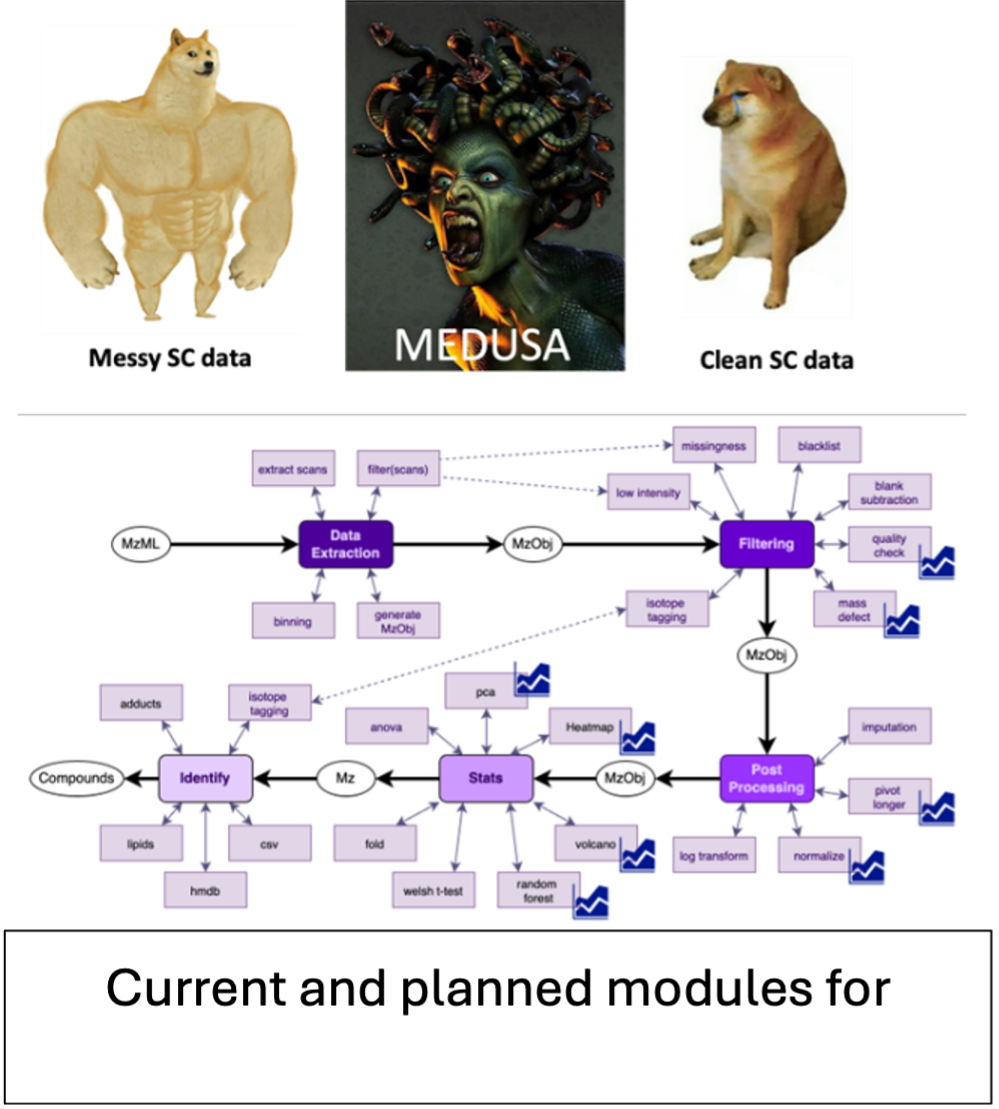
One Does Not Simply Process Single-Cell Data. Are you a developer looking for a quest worthy of song? We are recruiting brave programmers to help us refine MeDUSA, our cutting-edge tool for single-cell metabolomics. In the realm of biology, single cells are the "Unseen World". Their world is vast, chaotic, and holding secrets to drug resistance and cancer progression. Exploring this world requires tools of precision and immense power.
Our current weapon, while mighty, has grown heavy. We are looking for the Aragorns of R, the Gandalf’s of C++, and the Samwise Gamgees of Shiny to help us lighten the load and sharpen the blade. If you speak the language of code and have the courage to face messy, untargeted data, then speak friend and bid on us. We offer you the chance to leave your mark on a tool that pushes the boundaries of scientific discovery. What is MeDUSA?
MeDUSA (Metabolomics of Direct-Infusion Untargeted Single-cell Analysis) is a project for an R-based package with a GUI that specifically engineered to navigate the noisy, chaotic landscape of direct infusion mass spectrometry. Unlike traditional methods that rely on the structured order of chromatographic separation, MeDUSA operates in a "separation-free" environment, meaning it must act as a filter against the high noise and missing values inherent to single-cell measurements. Currently, the pipeline aims to manage the entire lifecycle of data: from the raw extraction of spectra to noise filtering, blank subtraction, and advanced statistical analysis like Random Forest and PCA.
The Quests (Proposed Project) A user-friendly GUI for researchers that utilizes the logic of MeDUSA functions to process and analyze single cell data.
Current status: Currently, the pipeline is spread over a docker container, and two GitHub repos. The code has been written by amateur scientific researchers. While the logic is sound, the code commented, from a programming point of view, it’s terrible :). A large portion of the workflow can only be ran in a docker module, while others have to be run independently off the docker environment. The code is unoptimized for large datasets.
The Mission; your task is as follows: 1. Refractor/ integrate / write the code to develop the data analysis platform after the “peak alignment processing step” 2. Ensure that the code/pipeline is modular, and can handle large datasets 3. Design a GUI to operate the workflow / package
Most importantly, you will have ultimate freedom in how you can achieve this, all ideas are welcome! The final product will be hosted online, and your contributions will be attributed on the website.
Resources: https://github.com/DirkWevers/GCTA https://github.com/laura-hetzel/MeDUSA/tree/main/MeDUSA
A user-friendly GUI for researchers that utilizes the logic of MeDUSA functions to process and analyze single cell data. 1. Refractor/ integrate / write the code to develop the data analysis platform after the “peak alignment processing step” 2. Ensure that the code/pipeline is modular, and can handle large datasets 3. Design a GUI to operate the workflow / package