Molecular Bookkeeping for structural analysis
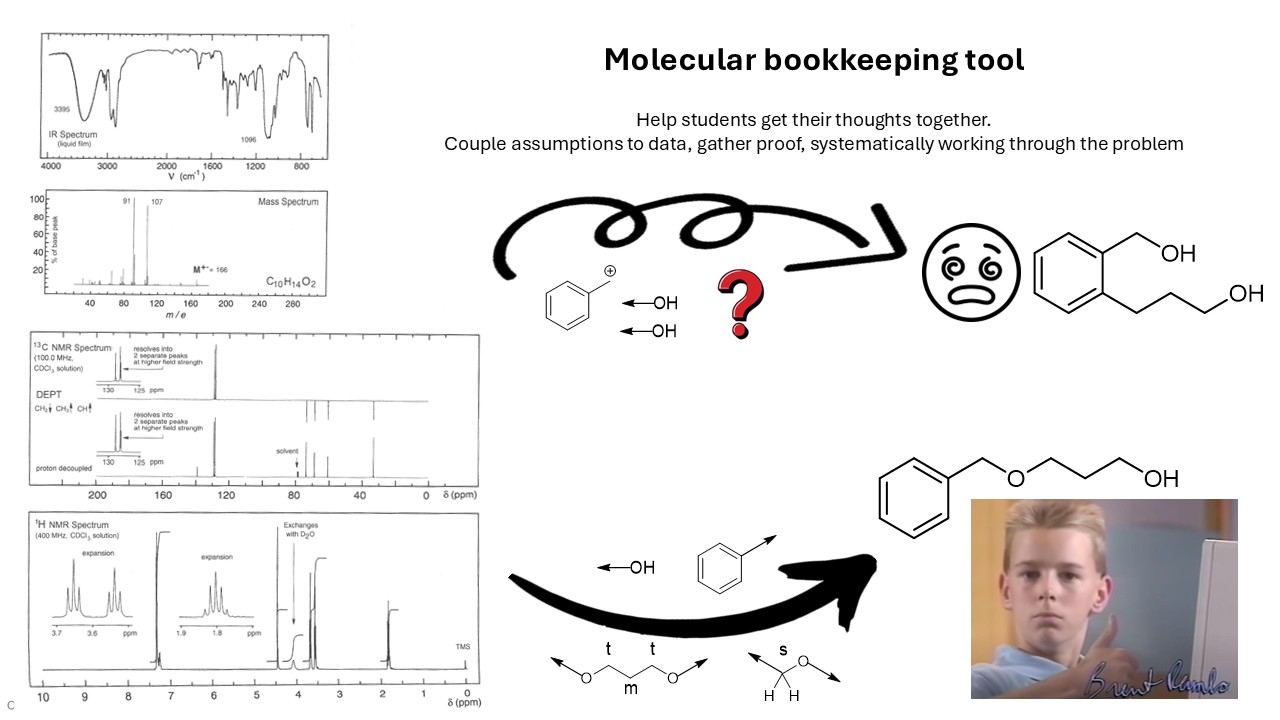
The aim of this project is to develop a piece of software which can help students learn how to work systematically on obtaining a chemical structure. The student is presented with a set of spectroscopic data and their goal is to provide the chemical structure. The approach may vary, but they have to rationalize the steps they make to avoid guessing and getting lost. The main challenge for students is not to get over- or underwhelmed and to tackle the problem at hand with a systematic approach. Students have to come up with molecular fragments, prove it is part of the molecule by relating the fragment to actual data, until all fragments have been found. The fragments then have to be assembled, prove it can or cannot be assembled in a certain way and then provide the end result. Students often get lost in keeping track of the data not yet assigned, or when insight changed, or things get assigned twice etc.
The goal for the software is to help students keep track of their analysis; a tool to do molecular bookkeeping. The software itself doesn't provide direct answers but only prompts when certain mistakes are made, like over-assignment of atoms. The origin of the mistake is for the student to figure out. The software helps the student to get into the habit of a systematic analysis, even when the order can differ every time.
Minimum viable: - New data can be added to an existing library (text format) - Data is displayed both textually/table and graphically (in a simple form), and can be zoomed in/out - A fragment database can be accessed and items selected, which can then be matched with datapoints. - Progression is tracked based on the matched data. When not everything is matched or data is matched twice, or another counter crosses a threshold, a warning is displayed. - in case of molecular symmetry, a datapoint can be matched with more than one fragment. Either an 'ignore' warning style work-around can be thought of, or the raw data can be appended to contain this information (to be discussed). - Selected/used fragments are displayed. - Matching can be (partially) undone, tracker is then also (partially) restored. - Fragments can be joined together by assigning wildcard-ID's in the textual version of the fragment (SMILES), fragments can have a pre-defined number of partners it can/need to match with. - The assembled end-result is a generated/combined SMILES string, which is then checked with the answer (provided with the raw textinput) - A manual check is implemented: a student can enter a CAS-number or SMILES string at any time to check if the end product is correct (answer is provided with raw text input)
"Fully" complete: - Data can be added as a compiled package (encrypted) form, and extracted by the software. Students can decide which packages to download from a place like brightspace. Teacher must be able to compile this from a set of textfiles, a seperate tool is required, or a separate tool within the software? - Graphically visualized includes Lorentzian line shape for individual datapoints. Alternatively, the actual real-spectrum can be shown (available as .svg file) - Graphs have hover-over highlighting, very useful for overlapping signals in the data. - Fragments coupled to data change in color to keep track of their status. - Several commands can be performed (calculation instructions, based on the information of the raw data) or checked. - Fragment database is visually rendered using SMILES, new fragments can be added by SMILES